In my last blog post we fitted a generalized linear model to count data using a Poisson error structure.
We found, however, that there was over-dispersion in the data – the variance was larger than the mean in our dependent variable.
Over-dispersion is a problem if the conditional variance (residual variance) is larger than the conditional mean. One way to check for and deal with over-dispersion is to run a quasi-poisson model, which fits an extra dispersion parameter to account for that extra variance.
Now let’s fit a quasi-Poisson model to the same data.
model2 <- glm(Students ~ Days, quasipoisson) summary(model2) Call: glm(formula = Students ~ Days, family = quasipoisson) Deviance Residuals: Min 1Q Median 3Q Max -2.00482 -0.85719 -0.09331 0.63969 1.73696 Coefficients: Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.990235 0.074789 26.61 <2e-16 ***
Days -0.017463 0.001539 -11.35 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for quasipoisson family taken to be 0.7939441)
Null deviance: 215.36 on 108 degrees of freedom
Residual deviance: 101.17 on 107 degrees of freedom
AIC: NA
Number of Fisher Scoring iterations: 5
The outcome of our attempt to account for over-dispersion is that the residual deviance has not changed.
The dispersion parameter, which was forced to be 1 in our last model, is allowed to be estimated here. In fact, it is estimated at .79.
This parameter tells us how many times larger the variance is than the mean. Since our dispersion was less than one, it turns out the conditional variance is actually smaller than the conditional mean. We have under-dispersion, not over.
We can extract the model coefficients in the usual way:
model2$coefficients (Intercept) Days 1.99023497 -0.01746317
Anyway – we now plot the regression. We set up a time axis running from 0 to 150 (the number of days). However, we include small increments of 0.1 in order to create a smooth appearance to our plot. We will evaluate the model on these values and then use those values to plot the model.
timeaxis <-seq 0="" 150="" 1="" pre="">
Anyway – we now plot the regression. We set up a time axis running from 0 to 150 (the number of days). However, we include small increments of 0.1 in order to create a smooth appearance to our plot. We will evaluate the model on these values and then use those values to plot the model.
timeaxis <-seq (0,150,0.1)
Now we use the predict() function to set up the fitted model values.
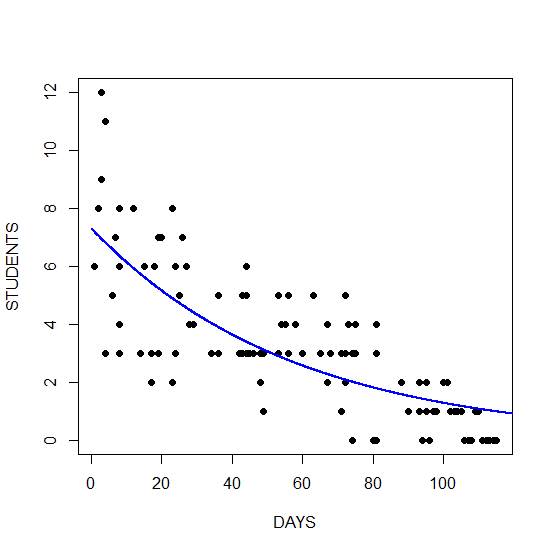
Y <- predict(model2, list(Days = timeaxis)) plot(Days, Number, xlab = "DAYS", ylab = "STUDENTS", pch = 16)
Finally, we plot the fitted model. We take the exponential of the fitted values because the fitted values are returned on a logarithmic scale. Taking the exponential back-transforms from the log scale to the original data.
lines(timeaxis, exp(Y), lwd = 2, col = "blue")
The graph shows a non-linear decrease in cases with number of days. Of course, instead of taking the exponential of the fitted values, we could also have used the predict() function together with the argument type = “response”.
Z <- predict(model2, list(Days = timeaxis), type = "response") plot(Days, Number, xlab = "DAYS", ylab = "NUMBER", pch = 16) lines(timeaxis, Z, lwd = 2, col = "red")
Let’s calculate the impact on the number of cases arising from a one day increase along the time axis. First we take the exponential of the coefficients.
coeffs <- exp(coef(model2)) coeffs (Intercept) Days 7.3172529 0.9826884
We calculate the 95% confidence interval (upper and lower confidence limits) as follows:
CI <- exp(confint.default(model2))
CI
2.5 % 97.5 %
(Intercept) 6.3195674 8.4724454
Days 0.9797296 0.9856562
We can calculate the change in number of students presenting with the disease for each additional day, as follows:
1 - 0.9826884 [1] 0.0173116
The reduction (rate ratio) is approximately 0.02 cases for each additional day.
****
See our full R Tutorial Series and other blog posts regarding R programming.
About the Author: David Lillis has taught R to many researchers and statisticians. His company, Sigma Statistics and Research Limited, provides both on-line instruction and face-to-face workshops on R, and coding services in R. David holds a doctorate in applied statistics.


Hi, I still have a question. Is it fine to apply a quasipoisson model to under dispersed data? Are there better ways to deal with underdispersion in R?
Thanks very much for the post. I would love to know how to use the Wald test to test for overdispersion in a Poisson and negative binomial regression model.
Thank you in advance
Hi,
Just wanted to say thank you SO much for all these posts. They really helped me to understand GLM and their purpose….especially since I have a final tomorrow 🙂
You really saved me.
Hi Am also playing with the possion and quasi poisson in glm.
I have found that the parameter fitting is identical using both families. It is only the dispersion parameter that changes.
Can anyone explain this?
That’s what quasi poisson is. It fits an extra parameter that allows the variance > mean. Poisson doesn’t.
great post! i just have want to underline that: the term quasipoisson in the formula of glm() is not a quasipoisson distribution. i here quote Zuur’s book pp.226(mixed model effects and their extensions in ecology)
quote: “specifiying the family option as quasipoisson instead of poisson gives the imporession that there is a quasi-Poisson distribution but there is no such thing! all we do here is specify the mean and variance relationchip and an exponential link between the expected values and explanatory variables. it is a software issue to call this ‘quasipoisson’. Do not write in your report or paper that you used a quasi-Poisson distribution.”
thus saying here that you used a quasipoisson is a mistake.
Hi Fabio, it wouldn’t be a mistake to say you ran a quasipoisson model, but you’re right, it is a mistake to say you ran a model with a quasipoisson distribution. The difference is subtle. As David points out the quasi poisson model runs a poisson model but adds a parameter to account for the overdispersion.
Karen
Thanks for this great post. When I use a quasi-poisson model to get the dispersion parameter for 8 different outcomes, I get values ranging from 1.24 – 2. What is a good “cutoff” for overdipsersion? Are all of these overdispersed since they are >1? Just trying to get a better sense of how to make this decision. Thanks!
Thanks for writing this helpful tutorial. I’m trying to recreate and am wondering where the “Number” variable come from in your first plot? Thanks!